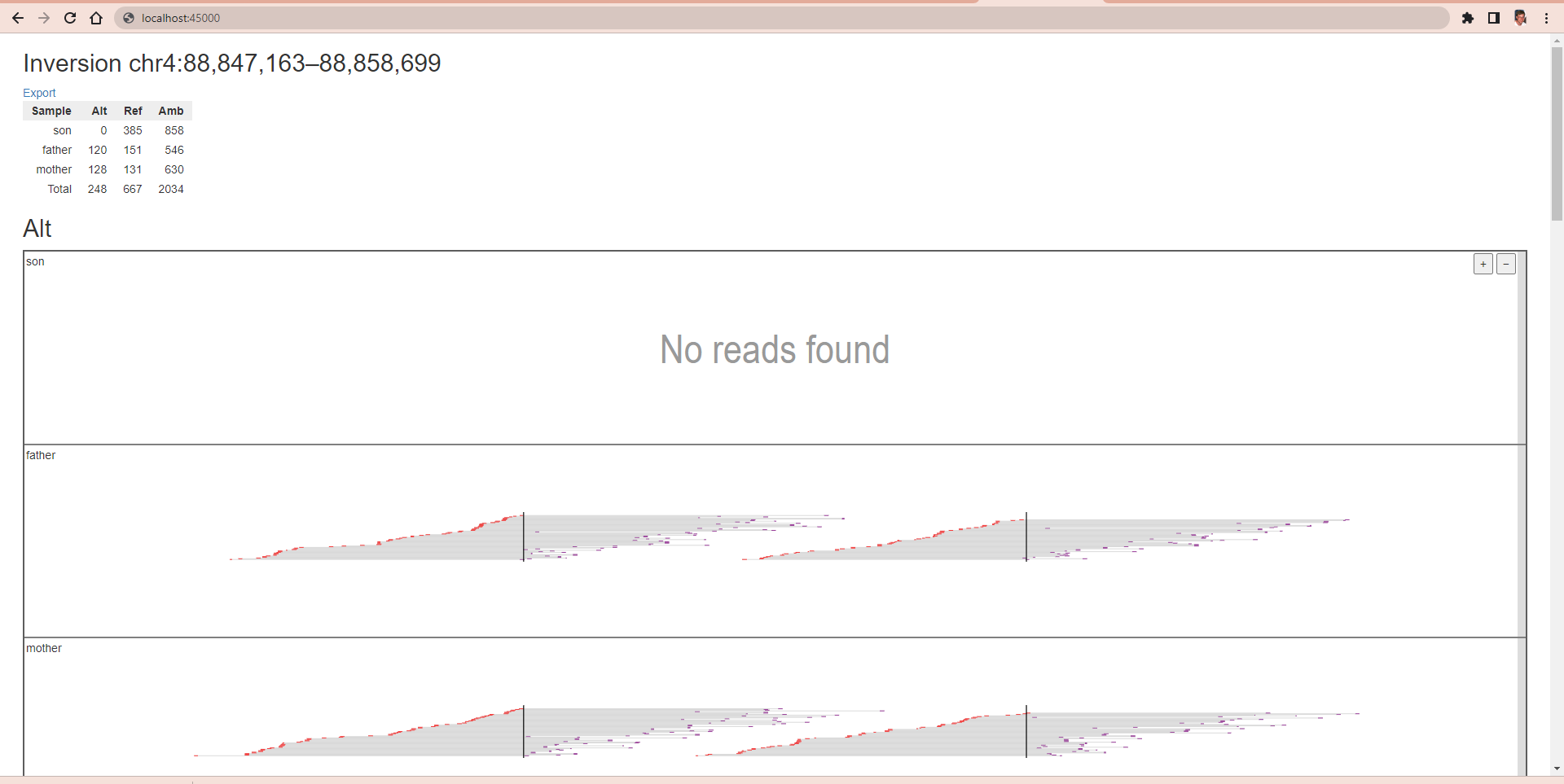
svviz visualizes high-throughput sequencing data relevant to a structural variant. Only reads supporting the variant or the reference allele will be shown. svviz can operate in both an interactive web browser view to closely inspect individual variants, or in batch mode, allowing multiple variants (annotated in a VCF file) to be analyzed simultaneously.
This application requires a interactive tunnel session
Allocate an interactive session and run the program.
Sample session (user input in bold):
[user@biowulf ~]$ sinteractive --gres=lscratch:10 --tunnel
salloc.exe: Pending job allocation 26710013
salloc.exe: job 26710013 queued and waiting for resources
salloc.exe: job 26710013 has been allocated resources
salloc.exe: Granted job allocation 26710013
salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn3094 are ready for job
Created 1 generic SSH tunnel(s) from this compute node to
biowulf for your use at port numbers defined
in the $PORTn ($PORT1, ...) environment variables.
Please create a SSH tunnel from your workstation to these ports on biowulf.
On Linux/MacOS, open a terminal and run:
ssh -L 45000:localhost:45000 biowulf.nih.gov
For Windows instructions, see https://hpc.nih.gov/docs/tunneling
[user@cn3094 ~]$ echo $PORT1
45000
[user@cn3094 ~]$ module load svviz
[user@cn3094 ~]$ svviz --port $PORT1 --type inv \
-b $SVVIZ_EXAMPLES/example1/son.sorted.bam \
-b $SVVIZ_EXAMPLES/example1/father.sorted.bam \
-b $SVVIZ_EXAMPLES/example1/mother.sorted.bam \
$SVVIZ_EXAMPLES/example1/chr4_short.fa chr4 88847163 88858699
...
Starting browser at http://127.0.0.1:45000/
* Serving Flask app "svviz.web" (lazy loading)
* Environment: production
WARNING: This is a development server. Do not use it in a production deployment.
Use a production WSGI server instead.
* Debug mode: off
Create an ssh tunnel using value of $PORT1 (mac/linux or windows).
Then open a web brower and load the URL localhost:#####, where ##### is the value of $PORT1.

If you want to run another svviz command, type the keystroke combination Control+C within the sinteractive tunnel session to escape and stop the svviz process. You can then run other svviz commands.
When you are completely finished with svviz and need to end the sinteractive tunnel session, type exit:
[user@cn3094 ~]$ exit salloc.exe: Relinquishing job allocation 26710013 [user@biowulf ~]$