
RStudio is an integrated development environment (IDE)for R. It includes a console, syntax-highlighting editor that supports direct code execution, as well as tools for plotting, history, debugging and workspace management.
This is RStudio Server, a browser-based interface very similar to the standard RStudio desktop environment. RStudio Server can be much more responsive and a generally-better experience when used remotely, especially over a VPN.
This application can be utilized interactively via the HPC OnDemand web interface.
We recommend using HPC OnDemand to access RStudio Server. This uses Open OnDemand to provide a convenient interface to launch and access RStudio sessions in interactive jobs without requiring complex SSH tunneling setup.
To access RStudio Server via Open OnDemand, you navigate to the
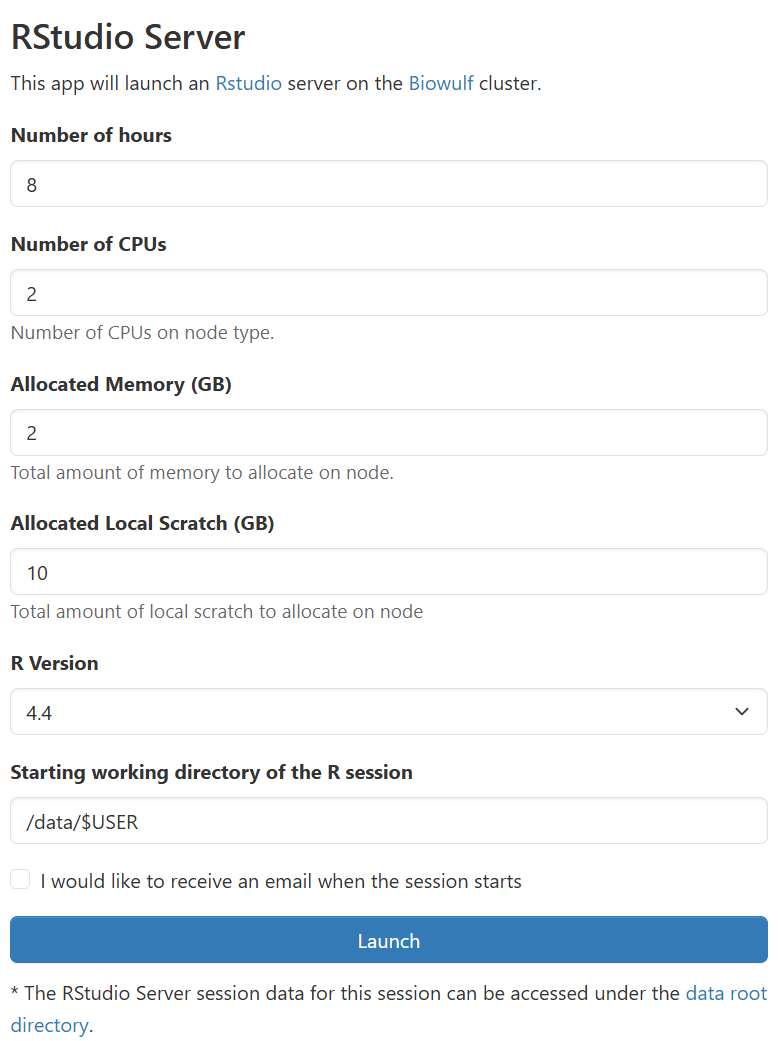
RStudio Server application page,
configure the resources your job requires, and launch your job. You may also select the startup "working directory",
equivalent to running cd before launching rstudio-server from the terminal.

For more information about Biowulf's Open OnDemand installation, access, and other services available please see our documentation.
Allocate an sinteractive session. It is required to allocate at least a small amount of lscratch for temporary storage for R.
It is also required to set up an sinteractive tunnel.
[user@biowulf ~]$ sinteractive --mem=10g --gres=lscratch:5 --tunnel
salloc.exe: Pending job allocation 15323416
salloc.exe: job 15323416 queued and waiting for resources
salloc.exe: job 15323416 has been allocated resources
salloc.exe: Granted job allocation 15323416salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn1640 are ready for job
Created 1 generic SSH tunnel(s) from this compute node to
biowulf for your use at port numbers defined
in the $PORTn ($PORT1, ...) environment variables.
Please create a SSH tunnel from your workstation to these ports on biowulf.
On Linux/MacOS, open a terminal and run:
ssh -L 39689:localhost:39689 user@biowulf.nih.gov
For Windows instructions, see https://hpc.nih.gov/docs/tunneling
[user@cn1640 ~]$ module load rstudio-server
[+] Loading gcc 9.2.0 ...
[-] Unloading gcc 9.2.0 ...
[+] Loading gcc 9.2.0 ...
[+] Loading openmpi 4.0.5 for GCC 9.2.0
[+] Loading ImageMagick 7.0.8 on cn4280
[+] Loading HDF5 1.10.4
[-] Unloading gcc 9.2.0 ...
[+] Loading gcc 9.2.0 ...
[+] Loading NetCDF 4.7.4_gcc9.2.0
[+] Loading pandoc 2.17.1.1 on cn4280
[+] Loading pcre2 10.21 ...
[+] Loading R 4.2.2
[+] Loading rstudio-server 2023.03.0-386
[user@cn1640 dir]$ rstudio-server
Please ensure you have set up the SSH port forwarding as described in the sinteractive instructions.
Please connect to http://localhost:39689/auth-sign-in?user=test2&password=nRmzfPWh_X8Z-03hbDjPz3bm
Use your username 'user' and the pasword 'nRmzfPWh_X8Z-03hbDjPz3bm' to login
If you have set up the SSH tunnel with the command or instructions provided (please make sure to use the specific port/command given by your interactive session) you should be able to load the link and have RStudio Server load in your browser.
If you are presented with a login screen, please use your username and the password provided