


|
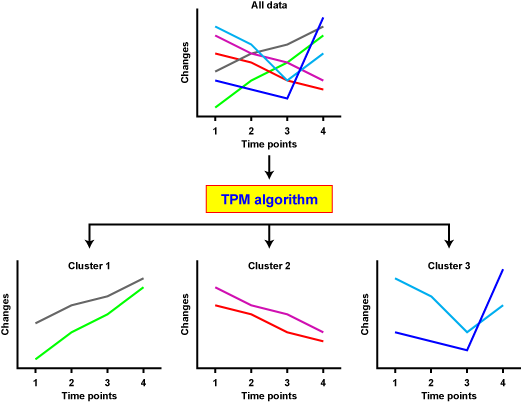
TPM algorithm clusters any time-series data set, specifically iTRAQ LC-MS/MS data sets. The data points that have a similar behavior over the time course are clustered together.
Download a standalone version of TPM: TPM.zipTPM algorithm was developed by Fahad Saeed at the Epithelial Systems Biology Laboratory, National Heart, Lung, and Blood Institute in Bethesda, Maryland USA. If you use this algorithm please cite the following paper: Fahad Saeed, Trairak Pisitkun, Mark Knepper and Jason D Hoffert, "Mining Temporal Patterns from iTRAQ Mass Spectrometry (LC-MS/MS) Data", The Proceedings of the ISCA 3rd International Conference on Bioinformatics and Computational Biology (BiCoB), Vol 1. pp 152-159 New Orleans, Louisiana, USA, March 23-25, 2011 (ISBN: 978-1-880843-81-9) Link to full paper: arXiv:1104.5510v1 If you have any question or want to report a bug about the algorithm please contact us. |
|
Input format
Data Input: The data is assumed to be the following. The first column is considered the peptide sequence or any other nomenclature that you are analyzing.
The columns that follow are the numbers that represent the entity that you wish to cluster for each time point (t1, t2, ? etc).
For example:
| peptide_1 | 0.223 | 0.440 | 0.232 | 0.232 |
| peptide_2 | 0.232 | 0.870 | 0.670 | 0.230 |
| peptide_3 | 0.670 | 0.440 | -0.340 | -0.220 |
| peptide_4 | 0.232 | 0.212 | 0.909 | 0.313 |
| peptide_5 | 0.771 | 0.111 | -0.313 | 0.111 |
The example above shows the tab-delimited text with the first column representing peptide names.
The second column represents the numbers for the first time point, the third column for the second time point and so on.
Please make sure that you don't have special characters ('"{}*^&!<>) in the input.
Disclaimer | Accessibility | CIT | NHLBI | NIH | DHHS | USA.gov