The NIH HPC group plans, manages and supports
high-performance computing systems specifically for use by the
intramural NIH community. These systems include
Biowulf,
a 105,000+ processor
Linux cluster;
Helix, an interactive system for
file transfer and management, and
Helixweb, which provides a number of web-based
scientific tools. We provide access to a wide range of computational
applications for genomics, molecular and structural biology, mathematical and
graphical analysis, image analysis, and other scientific fields.
The continued growth and support of NIH's Biowulf cluster is dependent upon
its demonstrable value to the NIH Intramural Research Program. If you publish
research that involved significant use of Biowulf, please cite the cluster.
Suggested citation text:
This work utilized the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov).
|
Quick Links
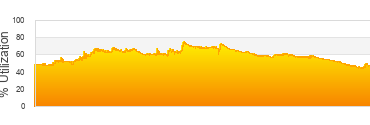
Biowulf Utilization
Sunday, January 25th, 2026
|
Last 24 hrs
|
|
39,412 jobs submitted
28,391 jobs completed
1,180,476 CPU hrs used
|
19 NIH Institutes
129 Principal Investigators
183 users
|
Announcements
|
Recent Papers that used Biowulf & HPC Resources
|
 A convenient single-cell assay for the newly synthesized transcriptome reveals FLI1 regulon downregulation during T cell activation
A convenient single-cell assay for the newly synthesized transcriptome reveals FLI1 regulon downregulation during T cell activation
Lyu, J; Xu, X; Chen, C; ,
Cell Rep Methods
, DOI://10.1016/j.crmeth.2025.101252 (2025)
A CGG Repeat Expansion in CSNK1E Associated with Progressive Myoclonic Epilepsy with Incomplete Penetrance
Akçimen, F; Alvarez Jerez, P; Guliyeva, U et al.
Mov Disord
, DOI://10.1002/mds.30326 (2025)
Excess CMS morbidity and FEMA declarations: Phenome Wide Association Study of Generalized Linear Model interactions between CMS diagnostic code utilization and FEMA Incident types, 1999-2020
Williams, N
Inform Med Unlocked
, DOI://10.1016/j.imu.2025.101716 (2025)
Artificial intelligence-based models for quantification of intra-pancreatic fat deposition and their clinical relevance: a systematic review of imaging studies
Joshi, T; Virostko, J; Petrov, MS; ,
Eur Radiol
, DOI://10.1007/s00330-025-11808-6 (2026)
 NAT10 promotes cancer metastasis by modulating p300/CBP activity through chromatin-associated tRNA
NAT10 promotes cancer metastasis by modulating p300/CBP activity through chromatin-associated tRNA
Amin, R; Ha, NH; Qiu, T et al.
Mol Cell
, DOI://10.1016/j.molcel.2025.11.010 (2025)
Epigenetic dysregulation in aged muscle stem cells drives mesenchymal progenitor expansion via IL-6 and Spp1 signaling
Riparini, G; Mackenzie, M; Naz, F et al.
Nat Aging
, DOI://10.1038/s43587-025-01002-0 (2025)
Response of iPSC-derived neurons from individuals with treatment-resistant depression to (2 R,6 R)-hydroxynorketamine and reelin: an exploratory study
Johnston, JN; Yuan, P; Kadriu, B et al.
Transl Psychiatry
, DOI://10.1038/s41398-025-03724-6 (2025)
 Interlocking host and viral cis-regulatory networks drive Merkel cell carcinoma
Interlocking host and viral cis-regulatory networks drive Merkel cell carcinoma
Miao, L; Milewski, D; Coxon, A et al.
J Clin Invest
, DOI://10.1172/JCI188924 (2025)
|
|
 All Services Operational
All Services Operational All Services Operational
All Services Operational